Introduction
Loess is a class of regression models that allows us to estimate the regression function by fitting a simple (and different) model to each point in the domain . This is done by paying attention to the points closest to a particular target point .
In the classic simple linear regression we have the following model:
where is:
and in the case of loess with approximate by a polynomial weigthed by a function that assigns higher weights to points that are closer to :
Procedure:
Let denote a set of values for a particular variable and let represent the corresponding response variable.
Find the closest points to the target point . Let’s denote this set
- For implementation, keep track of these points and their distances
Find the furthest distance among those points. Let’s denote this quantity as
Calculate the weight function for each using the tricube weight function:
where :
- Calculate the prediction by solving weigthed least squares. The normal equation can be expressed as followed:
You need to use only the closest points for each target point
- In the case of a polynomial of degree 1:
- W is a 2x2 diagonal matrix
- X is a kx2 matrix (column 1 it is a column of 1’s for the intercept)
- y is a kx1 vector
- In the case of a polynomial of degree 2:
- W is a 3x3 diagonal matrix
- X is a kx3 matrix
- y is a kx1 vector
- In the case of a polynomial of degree 1:
With the estimates of you can calculate
Implementation
In the implementation, I included an option to not use all the points as because when there are many samples, the fit takes long time.
Also, to solve the normal equation above, I did a QR decomposition for stability 1.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy.linalg import qr, pinv
from scipy.linalg import solve_triangular
def loess(X, y, alpha, deg, all_x = True, num_points = 100):
'''
Parameters
----------
X : numpy array 1D
Explanatory variable.
y : numpy array 1D
Response varible.
alpha : double
Proportion of the samples to include in local regression.
deg : int
Degree of the polynomial to fit. Option 1 or 2 only.
all_x : boolean, optional
Include all x points as target. The default is True.
num_points : int, optional
Number of points to include if all_x is false. The default is 100.
Returns
-------
y_hat : numpy array 1D
Y estimations at each focal point.
x_space : numpy array 1D
X range used to calculate each estimation of y.
'''
assert (deg == 1) or (deg == 2), "Deg has to be 1 or 2"
assert (alpha > 0) and (alpha <=1), "Alpha has to be between 0 and 1"
assert len(X) == len(y), "Length of X and y are different"
if all_x:
X_domain = X
else:
minX = min(X)
maxX = max(X)
X_domain = np.linspace(start=minX, stop=maxX, num=num_points)
n = len(X)
span = int(np.ceil(alpha * n))
#y_hat = np.zeros(n)
#x_space = np.zeros_like(X)
y_hat = np.zeros(len(X_domain))
x_space = np.zeros_like(X_domain)
for i, val in enumerate(X_domain):
#for i, val in enumerate(X):
distance = abs(X - val)
sorted_dist = np.sort(distance)
ind = np.argsort(distance)
Nx = X[ind[:span]]
Ny = y[ind[:span]]
delx0 = sorted_dist[span-1]
u = distance[ind[:span]] / delx0
w = (1 - u**3)**3
W = np.diag(w)
A = np.vander(Nx, N=1+deg)
V = np.matmul(np.matmul(A.T, W), A)
Y = np.matmul(np.matmul(A.T, W), Ny)
Q, R = qr(V)
p = solve_triangular(R, np.matmul(Q.T, Y))
#p = np.matmul(pinv(R), np.matmul(Q.T, Y))
#p = np.matmul(pinv(V), Y)
y_hat[i] = np.polyval(p, val)
x_space[i] = val
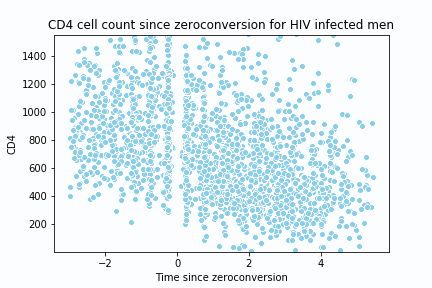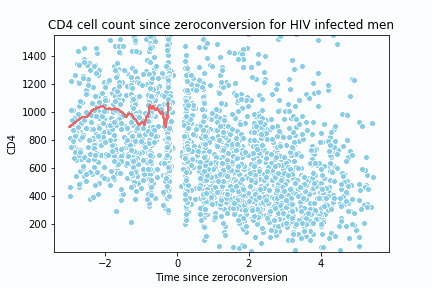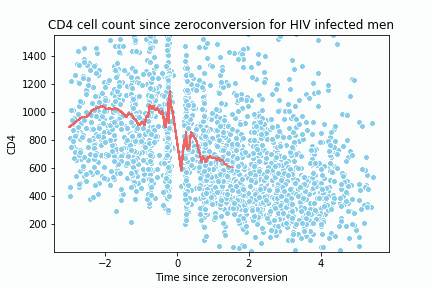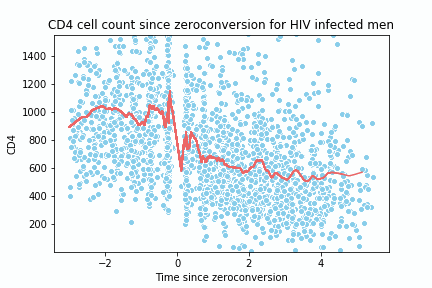
return y_hat, x_spaceTo test the implementation and create the animation, I replicate the following lecture notes 2. The dataset is located here 3.
import matplotlib.animation as animation
from matplotlib.animation import FuncAnimation, PillowWriter
c4 = pd.read_csv("cd4.data.txt", delim_whitespace=True, header=None)
y_hat2, x_space2 = loess(c4[0], c4[1], 0.05, 1, all_x = True, num_points = 200)
d = {'x': x_space2, 'y': y_hat2}
c4_pred = pd.DataFrame(data = d)
c4_pred = c4_pred.sort_values(by = ['x'])
c4_pred = c4_pred.reset_index(drop=True)
def animate(i):
x_data = c4_pred.x.values[:i]
y_data = c4_pred.y.values[:i]
#sns.scatterplot(c4[0], c4[1], color ='skyblue')
sns.lineplot(x= x_data, y=y_data, color = '#EE6666')
fig = plt.figure()
sns.scatterplot(c4[0], c4[1], color ='skyblue')
plt.ylim(1, 1550)
plt.xlabel("Time since zeroconversion")
plt.ylabel("CD4")
plt.title("CD4 cell count since zeroconversion for HIV infected men")
writer = animation.PillowWriter()
ani = FuncAnimation(fig, animate, frames = np.arange(0, 2376, 40))
ani.save('ani_cd4.gif', writer=writer)